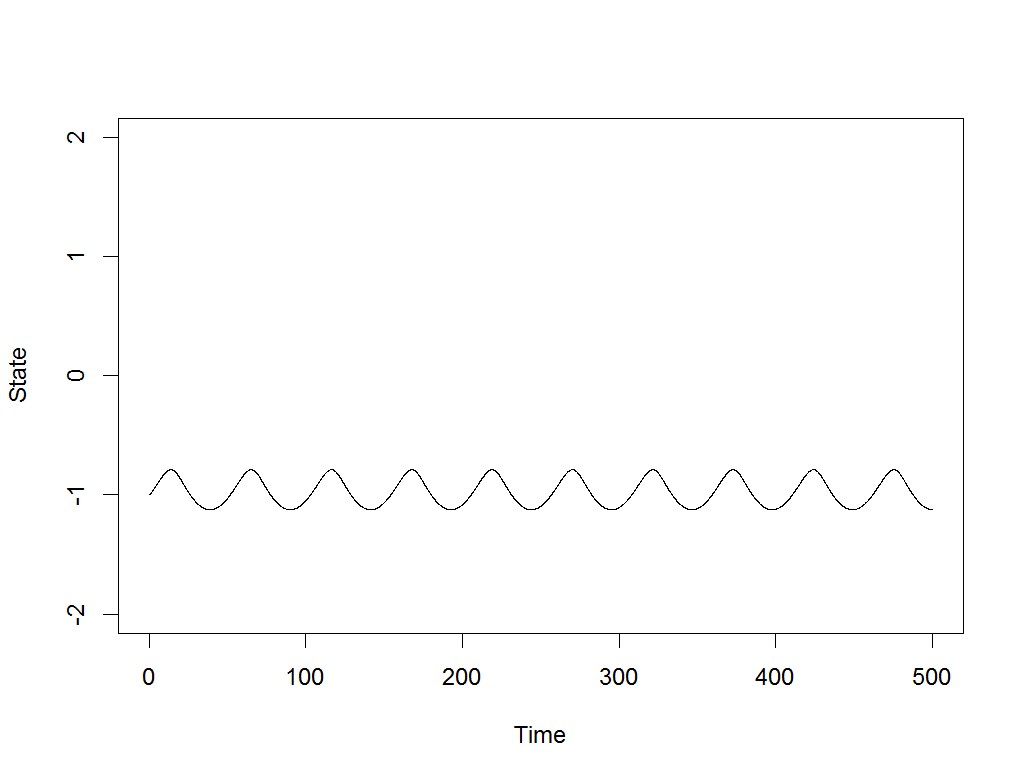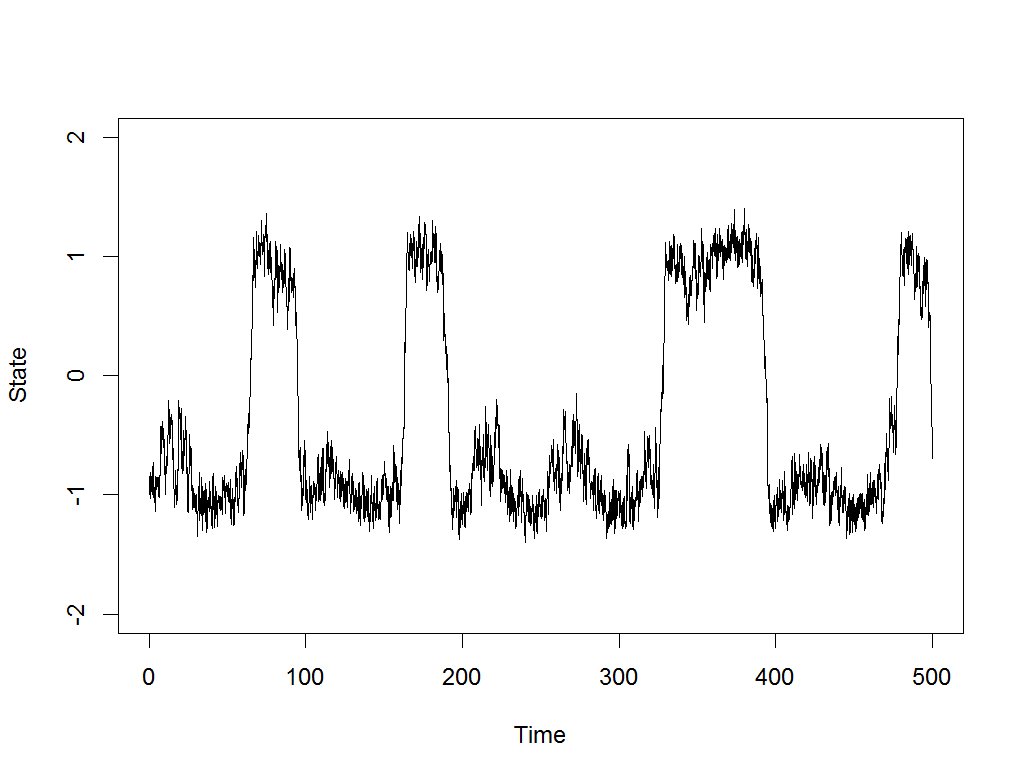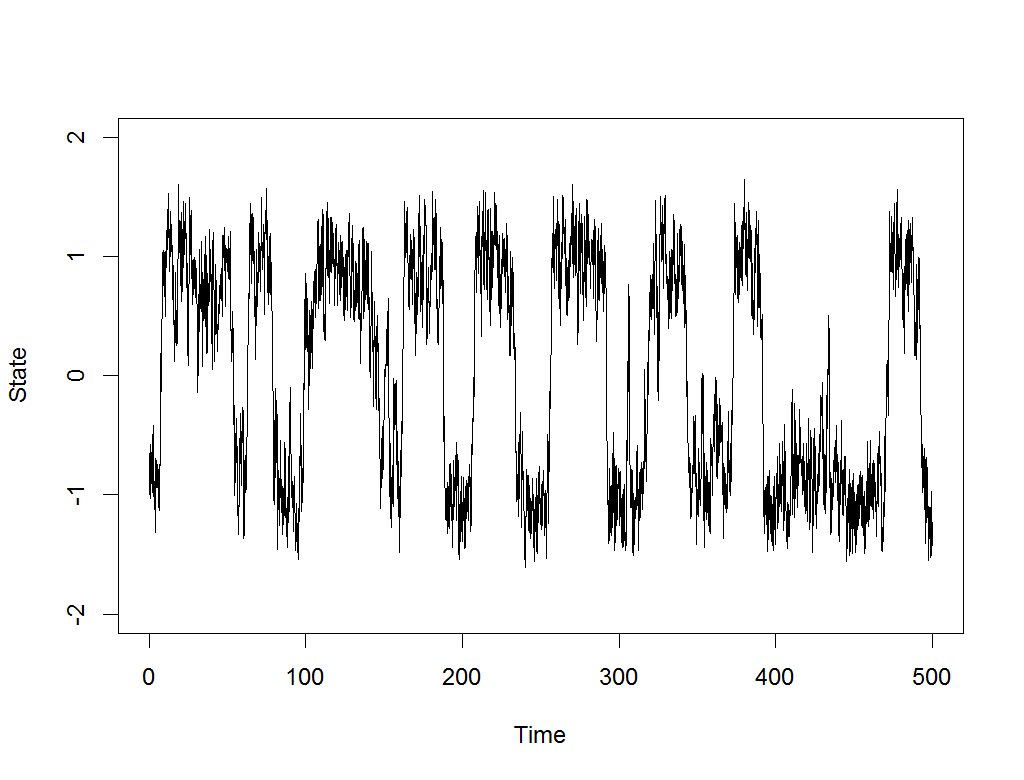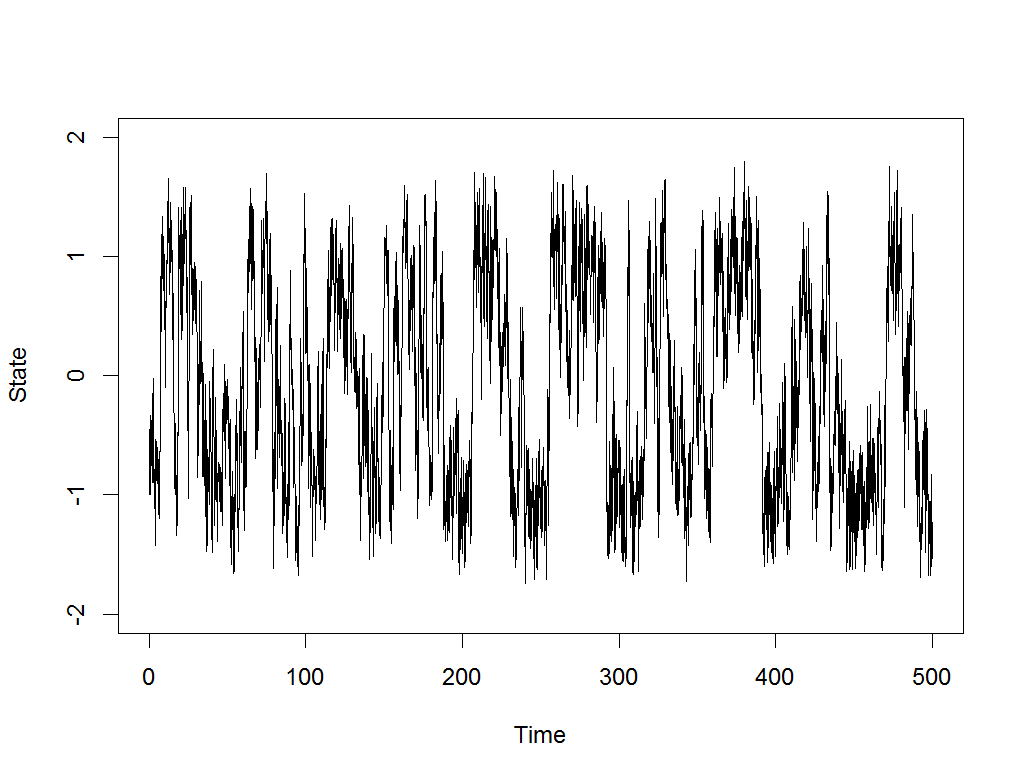
Exploration is the drum beat of any intelligent, self-improving system.
Key: Red: visited locations. Yellow: Last path taken. Cyan: Lowest-density frontier.
Most modern AI systems are built on machine learning, and machine learning achieves nothing without good, comprehensive data about its task. That’s why I’ve written a new paper, Meta-learning to Explore via Memory Density Feedback, demonstrating an improved approach to autonomous exploration. Watch above as the new RL-based agent, able to see only its own current coordinates and output movements, nonetheless learns to rapidly probe various maze problems, pushing its frontier and returning to lesser visited areas along the way.
[Read More]